| Tested Compounds | INHIBITORY CONCENTRATION VALUES (MIC/IC50) | Experimental Technique | 2D structure | PubChem CID |
|---|---|---|---|---|
| Azythromycin | MIC = 0.39 µg/mL | in vitro & in vivo |
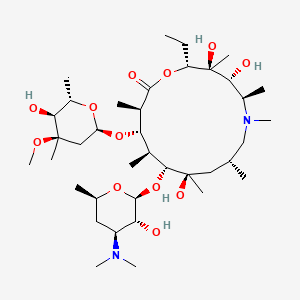
|
53477736 |
| Clarithromycin | MIC = 0.125 to 2 µg/mL | in vitro & in vivo |
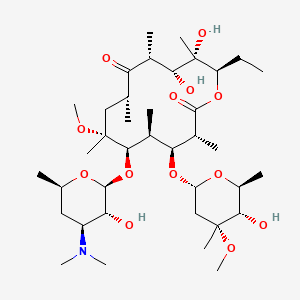
|
84029 |
| Dapsone | MIC = 0.04 µg/mL to 5 µg/mL | in vitro & in vivo |
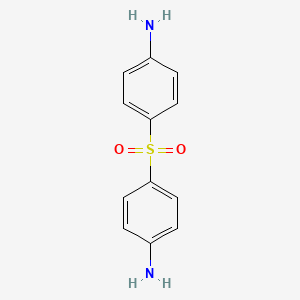
|
2955 |
| Clofazimine | MIC = 0.25 to 0.5 µg/ml | in vitro |
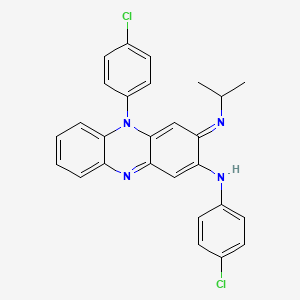
|
2794 |
| Rifampicin | MIC = 0.5 µ/ml | in vitro |
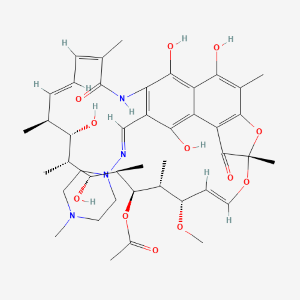
|
135398735 |
| Rifamycin | MIC = 0.06 to 0.125 µ/ml | in vitro |
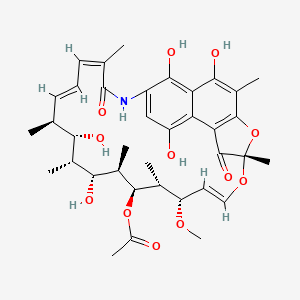
|
6324616 |
| Aminoglycosides - streptomycin | MIC = 0.25 µg/ml | in vitro |
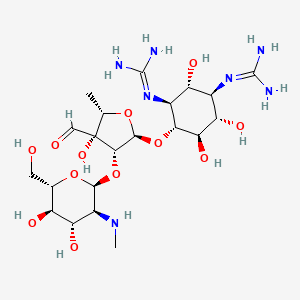
|
19649 |
| Ciprofloxacin | MIC = 1.15 µg/ml; 1–3 uM | in vitro |
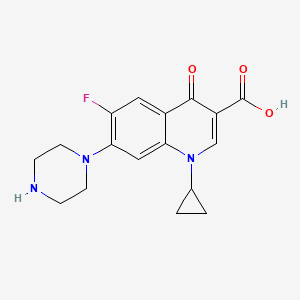
|
2764 |
| Telacebec (Q203) | MIC = 15 or 7.5 ng/mL | in vitro & in vivo |
.png)
|
68234908 |
| TB47 | MIC = 0.2 - 0.4µg/ml and > 50µg/ml | in vitro & in vivo |
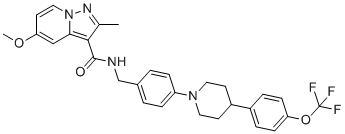
|
122188597 |
| Diazene-1,2-dicarboxamides | MIC = 5.67–7.25 µg/ml | in vitro |
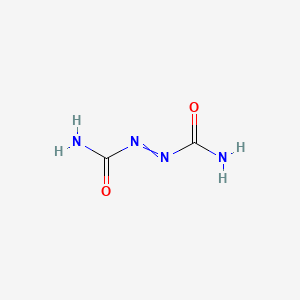
|
5462814 |
| Quinolinyl pyrimidines | MIC = Inactive | in vtro |

|
692596 |
| Pyrazolopyrimidines | MIC = 0.3–1.0 µM | in vtro |
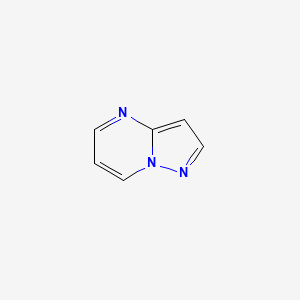
|
11636795 |
| Aminopyrazoles | MIC = 0.3–1.0 µM | in vtro |
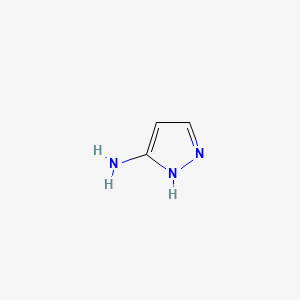
|
74561 |
| PA-824 | MIC = 13.1 µg/ml | in vitro |
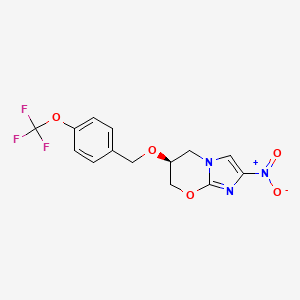
|
456199 |
| R207910 | MIC = 0.03 µg/ml | in vitro |
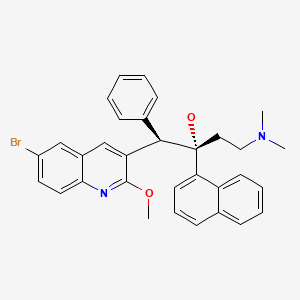
|
5388906 |
| KRM-1648 | MIC = 0.012 and 0.025 µg/ml | in vitro |
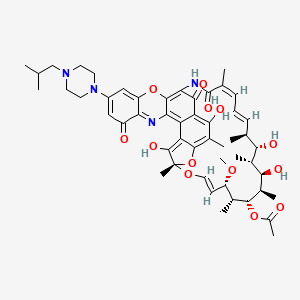
|
135431094 |
| SQ641 | MIC = 8 µg/ml | in vitro |
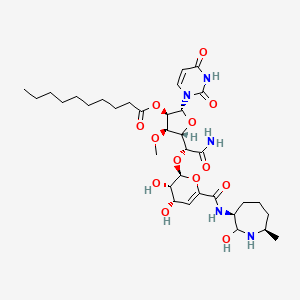
|
71589730 |
| Diarylthiazoles | MIC = 1 - 3 µM, 3 - 10 µM, > 10 µM | in vitro |

|
|
| 1,3-diaryltriazenes | IC50 = 2.82 and 0.06 µM | in vitro |
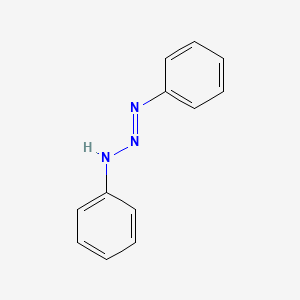
|
8689 |
| HMR 3004 | MIC = 5 – 40 µg/ml | in vitro |
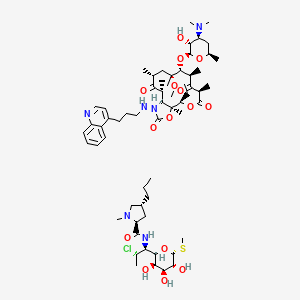
|
5481603 |
| HMR 3647 | MIC = 5 – 40 µg/ml | in vitro |
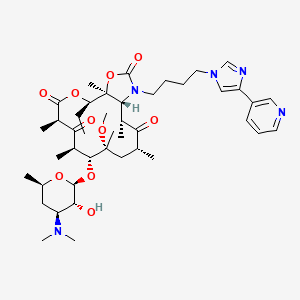
|
3002190 |
| Moxidectin | MIC = 4 – 32 µg/ml | in vitro |
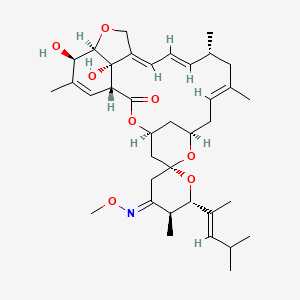
|
9832912 |
| Ivermectin | MIC = 4 – 32 µg/ml | in vitro |
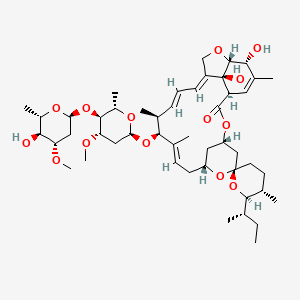
|
6321424 |
| Selamectin | MIC = 2 – 4 µg/ml | in vitro |
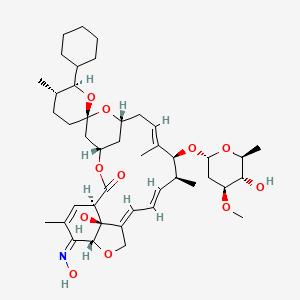
|
9578507 |
| Linezolid | MIC = 3 – 10 µM; 0.73 µg/ml | in vitro |
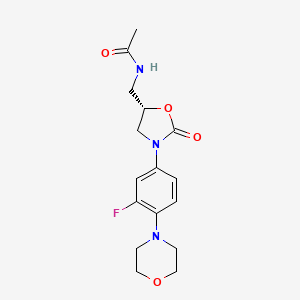
|
441401 |
| Prulifloxacin | MIC = 1 – 3 µM | in vitro |
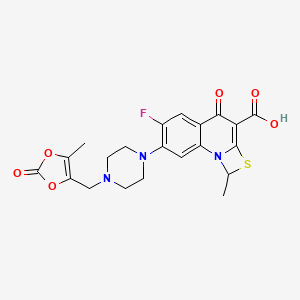
|
65947 |
| Sitafloxacin | MIC = 0.125 – 0.5 µg/ml | in vitro |
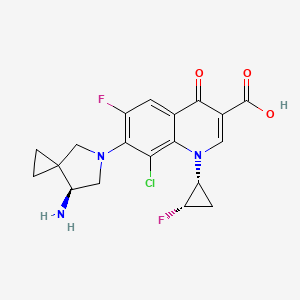
|
461399 |
| Moxifloxacin | MIC = 0.14 µg/ml | in vitro |
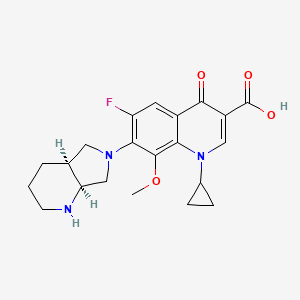
|
152946 |
| Sparfloxacin | MIC = 0.25 mg/l | in vitro |
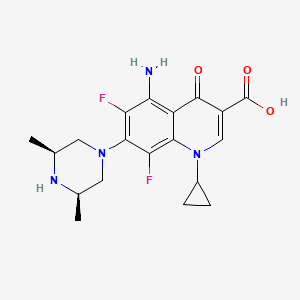
|
60464 |
| Amikacin | MIC = 0.5 – 0.65 µg/ml | in vitro |
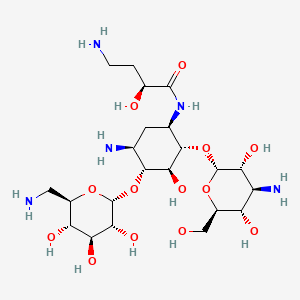
|
|
| ZINC38143792 (Euscaphic acid) | MIC = 50 µg mL−1 | in silico |
.png)
|
471426 |
| ZINC95485880 ( hydroxy-(hydroxymethyl)-dimethyl-BLAHone) | in silico |
-dimethyl-BLAHone).png)
|
||
| ZINC95486305 ( [(E)-5-[(2S,8S,9S,10S,13S,14R,16R,17S)-2,16-dihydroxy-4,4,9,13,14-pentamethyl-3,11-dioxo-2,7,8,10,12) | in silico |
-5-[(2S,8S,9S,10S,13S,14R,16R,17S)-2,16-dihydroxy-4,4,9,13,14-pentamethyl-3,11-dioxo-2,7,8,10,12).png)
|
||
| ZINC000018185774 (luteolin) | in silico |
.png)
|
||
| ZINC000095485921 ( 1,4,8-trihydroxy-5-(3-methylbut-2-enyl)xanthen-9-one) | in silico |
xanthen-9-one).png)
|
||
| ZINC000014417338 (Alpinumisoflavone) | in silico |
.png)
|
5490139 | |
| ZINC000005357841 ((6-methoxybenzo[1,3]dioxol-5-yl)BLAHone) | in silico |
![2D Structure of ZINC000005357841 ((6-methoxybenzo[1,3]dioxol-5-yl)BLAHone)](/images/testCompound2d/ZINC000005357841 ((6-methoxybenzo[1,3]dioxol-5-yl)BLAHone).png)
|
3781358 40529059 | |
| Phenothiazines | MIC = Inactive | in vitro |
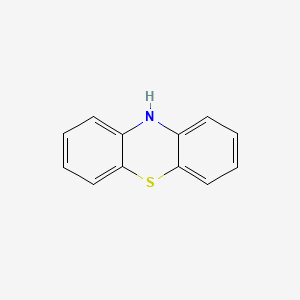
|
7108 |
| hydroxyquinolones | MIC = 1 – 3 µM | in vitro |
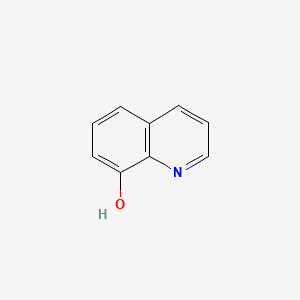
|
1923 |
| GyrB_Pyrollamide PubChem_25223515 | MIC = 0.3 – 1.0 µM | in vitro |
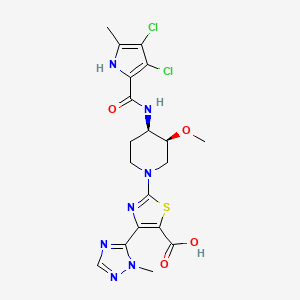
|
25223515 |
| GYRA-NTBI PubChem_15983305 2-oxo-1-[2-[4-[(3-oxo-4H-pyrido[3,2-b][1,4]oxazin-6-yl)methylamino]piperidin-1-yl]ethyl]quinoline-7-carbonitrile | MIC = Inactive | in vitro |
![2D Structure of GYRA-NTBI PubChem_15983305 2-oxo-1-[2-[4-[(3-oxo-4H-pyrido[3,2-b][1,4]oxazin-6-yl)methylamino]piperidin-1-yl]ethyl]quinoline-7-carbonitrile](/images/testCompound2d/GYRA-NTBI PubChem_15983305 2-oxo-1-[2-[4-[(3-oxo-4H-pyrido[3,2-b][1,4]oxazin-6-yl)methylamino]piperidin-1-yl]ethyl]quinoline-7-carbonitrile.png)
|
|
| GyrA_NTBI-analog | MIC = Inactive | in vitro |

|
|
| Ofloxacin | MIC = 1.26 – 2.0 µg/ml | in vitro |
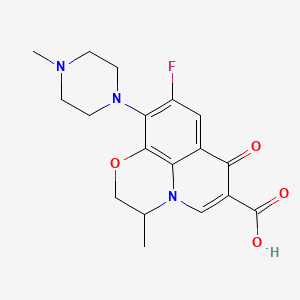
|
4583 |
| Doxycycline | MIC = Inactive | in vitro |
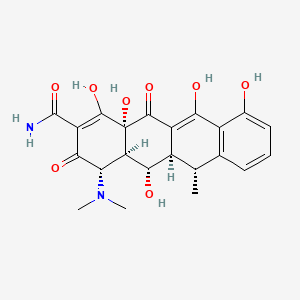
|
54671203 |
| Rifapentine | MIC = 0.125 – 0.5 µg/ml | in vitro |
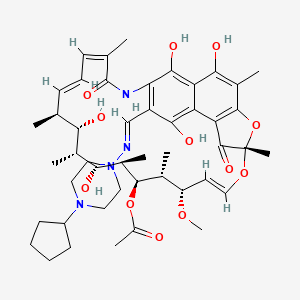
|
135403821 |
| Rifabutin | MIC = 0.1 – 0.4 µg/ml | in vitro |
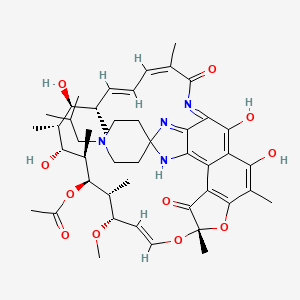
|
135398743 |
| 2,3,6-trihydroxymethyl benzoate (Sorindeia juglandifolia) | MIC = 62.5 µg/ml | in vitro |
.png)
|
|
| holadysamine (Holarrhena floribunda) | MIC = 50 µg/ml | in vitro |
.png)
|
132602823 |
| holaphyllinol (Holarrhena floribunda) | MIC = 125 µg/ml | in vitro |
.png)
|
132602821 |
| holamine or holaphyllamine (Holarrhena floribunda) | MIC = 125 µg/ml | in vitro |
.png)
|
12310548 |